How to deal with the "Aborting commit .... remains in conflict error"
% svn ci
svn: Commit failed (details follow):
svn: Aborting commit: './xmpl_rv' remains in conflict
% svn resolved xmpl_rv
Resolved conflicted state of 'xmpl_rv'
% svn ci
Wednesday, June 30, 2010
Monday, June 28, 2010
Reconstruction for Alexia
I'm back in Berkeley and very grateful to be in the cool weather and sleeping in my own bed. I seriously don't know how East Coast people handle that summer humidity.
I've spent the day getting my reconstruction code into shape for passing it over to Alexia to debug. Some lessons learned today are as follows:
1) I need to stop doing this "copy and paste" thing with python. Time to write up some more functions and get everything in execution mode... Alexia is right, I am wrong.
2) Some plotting tricks:
I've turned my plotting code into some plotting functions (see plotWPS and plotXi functions ../pythonjess/correlationData.py).
These functions now save the plots into the runDirectory as png files. You can look at them over terminal by typing:
I've spent the day getting my reconstruction code into shape for passing it over to Alexia to debug. Some lessons learned today are as follows:
1) I need to stop doing this "copy and paste" thing with python. Time to write up some more functions and get everything in execution mode... Alexia is right, I am wrong.
2) Some plotting tricks:
I've turned my plotting code into some plotting functions (see plotWPS and plotXi functions ../pythonjess/correlationData.py).
These functions now save the plots into the runDirectory as png files. You can look at them over terminal by typing:
> display plotname.png
They look like this (well why are white on black,
I need to figure out how to make them be white-on-black)
I need to figure out how to make them be white-on-black)
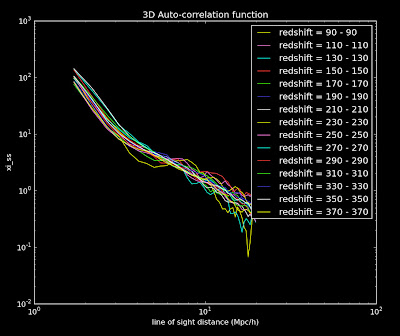
If you look at the inner working of these functions I use some commands do the pylab.gcf to set the size and print to a file. I think somewhere in there I can set up a color scheme. Should look into this. I used this web page to help me configure the above plot though.
3) I still don't really understand the different types of ways to write python functions. I've been defining functions as follows:
3) I still don't really understand the different types of ways to write python functions. I've been defining functions as follows:
def function(arguments):
.
.
----Insert function code here
.
.
----return(thing to return)
However it seems like I can also just have a set of code (like a main function in C) that calls a bunch of other functions and isn't a self-contained function... and Demitri said that I just need the following line at the top of the code to make it executable in python:
#!/usr/bin/python
But then at scicoder, Demitri also said that I should create everything as a class. So I guess I just need to figure out what the difference between a class, def, and then just code in a file is.... Maybe Demitri, Alexia, Josh or Adam can help me?
#!/usr/bin/python
But then at scicoder, Demitri also said that I should create everything as a class. So I guess I just need to figure out what the difference between a class, def, and then just code in a file is.... Maybe Demitri, Alexia, Josh or Adam can help me?
However, as a result of all this effort, I now have code that runs the reconstruction from head-to-tail, and re-runs it once you have already made the correlation functions, and re-runs it with different phi-binning. They are in the following files:
../pythonjess/jessreconstructionFull.py
../pythonjess/jessreconstruction.py
../pythonjess/rerunjessreconstruction.py
These files are the logfiles for today.
Best reconstruction Plot:
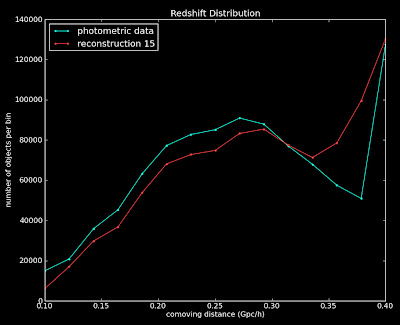
=
Labels:
Adam Pauls,
Alexia Schulz,
coding,
Demitri Muna,
Josh Meyers,
organization,
plotting,
reconstruction
Friday, June 25, 2010
Coding Wisdom from David Hogg
David Hogg came Scicoder to talk to us about Test-Driven Coding. He is pretty awesome.
Testing Your Code
1 bug / 1000 lines of code.
Ways to deal with this:
1) Test Driving Programming
2) Open-Source
3) Do Science with Code
Extreme Coding
1) Pair coding (coding together)
2) Stand up meetings (don't talk -- do)
3) Test Driving Programming (see below)
4) Minimal Implementation (re-factor frequently) only code things that you are actually going to use.
Test-Driven Programming
1) First write a test function to perform all possible tests you can imagine for your function.
2) Then run write the function and see how many of the tests it passes...
3) Keep modifying your function until it passes all the tests
4) Can't use code, check into repository unless it passes all tests
Functional Testing
1) Generate fake data + garbage
2) Run code
3) Did you get back what you put in?
4) Put assert statements in your code, such that if these assertions fail your code fails and spits out an error.
Have to be careful not to generate fake data that covered the entire spectrum of properties of your real data --- thus you could not be testing all cases or making all possible assertions.
But at the end of the day... what we really care about it this:
How do I know that my results are correct? ← (Probably not answerable)
or in real life...
Why do I think that my students results are correct?
- You do science with real data.
- You get results that you think you can publish and people will believe you (seriously this is what he said).
Blanton et. al. Luminosity Function Paper was originally a test by Mike Blanton to see if the SDSS data software pipeline was working and turned into one of the highest refereed papers out of the Sloan Survey. Thus testing really does matter.
On a side note, a link to my blog made it into Hogg's blog, and the thus circle is now complete.
Testing Your Code
1 bug / 1000 lines of code.
Ways to deal with this:
1) Test Driving Programming
2) Open-Source
3) Do Science with Code
Extreme Coding
1) Pair coding (coding together)
2) Stand up meetings (don't talk -- do)
3) Test Driving Programming (see below)
4) Minimal Implementation (re-factor frequently) only code things that you are actually going to use.
Test-Driven Programming
1) First write a test function to perform all possible tests you can imagine for your function.
2) Then run write the function and see how many of the tests it passes...
3) Keep modifying your function until it passes all the tests
4) Can't use code, check into repository unless it passes all tests
Functional Testing
1) Generate fake data + garbage
2) Run code
3) Did you get back what you put in?
4) Put assert statements in your code, such that if these assertions fail your code fails and spits out an error.
Have to be careful not to generate fake data that covered the entire spectrum of properties of your real data --- thus you could not be testing all cases or making all possible assertions.
But at the end of the day... what we really care about it this:
How do I know that my results are correct? ← (Probably not answerable)
or in real life...
Why do I think that my students results are correct?
- You do science with real data.
- You get results that you think you can publish and people will believe you (seriously this is what he said).
Blanton et. al. Luminosity Function Paper was originally a test by Mike Blanton to see if the SDSS data software pipeline was working and turned into one of the highest refereed papers out of the Sloan Survey. Thus testing really does matter.
On a side note, a link to my blog made it into Hogg's blog, and the thus circle is now complete.
Labels:
coding,
David Hogg,
extreme coding,
scicoder,
testing
Regular Expressions
Useful information about regular expressions. Reproduced from Ben Weaver's talk at Scicoder. Pdf is on repository here.
. stands for any one character
d.g matches dog, dg, dfg ....
[] matches a set of characters
d[aeiou]g matches dag, deg, dig, dog, dug but not dfg.
d[a-z] matches dag - dzg
Quantifiers
? = 0 or 1 {0,1}
+ = 1 or more {1,}
* = 0 or more {0,}
{i,j} = at least i, up to j
{i,} at least i, up to infinity
Examples....
do+g matches dog, doog, dooog, . . .
do*g matches dg, dog, doog, dooog, . . .
do?g matches dg or dog, nothing else.
do{2,3}g matches doog or dooog.
<.*> matches the entirety <h1>Title</h1>
<.*?> matches only <h1> or </h1> in <h1>Title</h1>
The anti-greed operation ? can be applied to any
quantifier.
.* is very, very greedy. Use it with caution.
<[a-z0-9/ ]+> might be better to use here
| is the symbol for ‘or’
dog|cat matches dog and cat.
Does not match dogat or docat (has low precedence)
( ) defines a group
Does not match anything on its own
do(g|c)at matches dogat or docat
^ matches the start of a string
^[A-Z] matches My dog has no nose.
^dog does not match
Note: different from [^ ]
$ matches the end of a string
dogs$ matches cats and dogs
The ‘end’ can (usually) be thought of as squeezed in
between the last character and the newline.
\ is the escape character
Turns off the special meanings of other metacharacters
\[0-9\] matches [0-9]
Also turns ordinary characters into metacharacters
\d = [0-9]
\D = [^0-9]
\s matches whitespace (space, tab, . . . )
\S matches non-whitespace
\w matches ‘alphanumeric’ = [A-Za-z0-9_]
\W = [^A-Za-z0-9_]
Python implements RE through a module, re
Differs from perl, which has RE built into the language
You don’t have to use re if you don’t need to
However, expressing an RE gets trickier
import re
RE must be distinguished from ordinary strings
’\b’ is the bell character
r’\b’ is a backslash followed by a b
Example:
Problem: Remove trailing comments plus any trailing
whitespace from a line.
line = ’keyword value \t # A keyword-value pair’
The re way:
commentRe = re.compile(r’^([^#]*?)(\s*)(#.*)?$’) # Reusable!
clean = commentRe.sub(r’\1’,line)
The string way:
try:
clean = line[0:line.index(’#’)].strip()
except ValueError: # if the string contains no comment
clean = line.strip()
For more look at the pdf.
. stands for any one character
d.g matches dog, dg, dfg ....
[] matches a set of characters
d[aeiou]g matches dag, deg, dig, dog, dug but not dfg.
d[a-z] matches dag - dzg
Quantifiers
? = 0 or 1 {0,1}
+ = 1 or more {1,}
* = 0 or more {0,}
{i,j} = at least i, up to j
{i,} at least i, up to infinity
Examples....
do+g matches dog, doog, dooog, . . .
do*g matches dg, dog, doog, dooog, . . .
do?g matches dg or dog, nothing else.
do{2,3}g matches doog or dooog.
<.*> matches the entirety <h1>Title</h1>
<.*?> matches only <h1> or </h1> in <h1>Title</h1>
The anti-greed operation ? can be applied to any
quantifier.
.* is very, very greedy. Use it with caution.
<[a-z0-9/ ]+> might be better to use here
| is the symbol for ‘or’
dog|cat matches dog and cat.
Does not match dogat or docat (has low precedence)
( ) defines a group
Does not match anything on its own
do(g|c)at matches dogat or docat
^ matches the start of a string
^[A-Z] matches My dog has no nose.
^dog does not match
Note: different from [^ ]
$ matches the end of a string
dogs$ matches cats and dogs
The ‘end’ can (usually) be thought of as squeezed in
between the last character and the newline.
\ is the escape character
Turns off the special meanings of other metacharacters
\[0-9\] matches [0-9]
Also turns ordinary characters into metacharacters
\d = [0-9]
\D = [^0-9]
\s matches whitespace (space, tab, . . . )
\S matches non-whitespace
\w matches ‘alphanumeric’ = [A-Za-z0-9_]
\W = [^A-Za-z0-9_]
Python implements RE through a module, re
Differs from perl, which has RE built into the language
You don’t have to use re if you don’t need to
However, expressing an RE gets trickier
import re
RE must be distinguished from ordinary strings
’\b’ is the bell character
r’\b’ is a backslash followed by a b
Example:
Problem: Remove trailing comments plus any trailing
whitespace from a line.
line = ’keyword value \t # A keyword-value pair’
The re way:
commentRe = re.compile(r’^([^#]*?)(\s*)(#.*)?$’) # Reusable!
clean = commentRe.sub(r’\1’,line)
The string way:
try:
clean = line[0:line.index(’#’)].strip()
except ValueError: # if the string contains no comment
clean = line.strip()
For more look at the pdf.
Thursday, June 24, 2010
SQL Commands (for accessing database)
Here is the database schema:
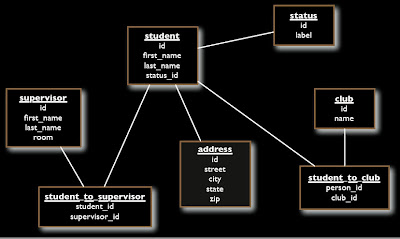
Below is for the database in the respository ../Database/Student database solution....
To start program:
cd
sqlite3 student_data.sqlite
Commands:
sqlite> .schema
CREATE TABLE "Club" ("id" INTEGER PRIMARY KEY NOT NULL ,"name" TEXT NOT NULL );
CREATE TABLE "Status" ("id" INTEGER PRIMARY KEY NOT NULL ,"label" TEXT NOT NULL );
CREATE TABLE "Student" ("id" INTEGER PRIMARY KEY NOT NULL ,"first_name" TEXT NOT NULL ,"last_name" TEXT NOT NULL , "status_id" INTEGER NOT NULL DEFAULT 0);
CREATE TABLE "student_to_club" ("student_id" INTEGER NOT NULL , "club_id" INTEGER NOT NULL , PRIMARY KEY ("student_id", "club_id"));
CREATE TABLE "student_to_supervisor" ("student_id" INTEGER NOT NULL , "supervisor_id" INTEGER NOT NULL , PRIMARY KEY ("student_id", "supervisor_id"));
CREATE TABLE "supervisor" ("id" INTEGER PRIMARY KEY NOT NULL ,"name" TEXT NOT NULL ,"room" TEXT NOT NULL );
sqlite> select count(*) FROM student;
100
sqlite> select count(first_name) FROM student;
100
sqlite> select count() FROM student;
100
sqlite> SELECT * FROM student LIMIT 5;
1|Cara|Rogers|1
2|Ori|Mejia|2
3|Leandra|Stevens|3
4|Danielle|Moody|1
5|Josiah|Barber|1
sqlite> SELECT first_name FROM student LIMIT 5;
Cara
Ori
Leandra
Danielle
Josiah
sqlite> SELECT first_name FROM student ORDER BY first_name ASC LIMIT 5;
Adara
Aileen
Alfreda
Amaya
Amber
sqlite> SELECT first_name FROM student ORDER BY first_name DESC LIMIT 5;
Yeo
Xantha
Wing
Wade
Timothy
sqlite> SELECT first_name, last_name FROM student ORDER BY last_name DESC LIMIT 5;
Galena|Zimmerman
Aileen|Wilkinson
Josephine|Wilkinson
Nerea|Whitney
Elmo|Webb
sqlite> SELECT first_name, last_name FROM student WHERE id=5;
Josiah|Barber
sqlite> SELECT first_name, last_name FROM student WHERE first_name like 'F%';
Fritz|Mccormick
Florence|Lang
sqlite> SELECT id, first_name, last_name FROM student WHERE id BETWEEN 10 and 15;
10|Leroy|Kent
11|Sandra|Carrillo
12|Raya|Thompson
13|Jael|Craig
14|Joshua|Forbes
15|Eve|Hinton
sqlite> SELECT id, first_name, last_name FROM student WHERE id in (15, 23, 6, 56, 9);
6|Wing|Gordon
9|Libby|Osborn
15|Eve|Hinton
23|Magee|Petersen
56|Philip|Parks
sqlite> SELECT id, first_name, last_name FROM student WHERE first_name in ('Libby', 'Philip');
9|Libby|Osborn
56|Philip|Parks
sqlite> SELECT sum(id) FROM student;
5050
sqlite> SELECT min(id) FROM student;
1
sqlite> SELECT max(id) FROM student;
100
sqlite> SELECT first_name, last_name, label FROM student JOIN status ON student.status_id = status.id LIMIT 5;
Cara|Rogers|Sophomore
Ori|Mejia|Senior
Leandra|Stevens|Freshman
Danielle|Moody|Sophomore
Josiah|Barber|Sophomore
sqlite> SELECT first_name, last_name, name FROM student JOIN student_to_club ON student_to_club.student_id = student.id JOIN club ON student_to_club.club_id = club.id LIMIT 5;
Cara|Rogers|Chess
Cara|Rogers|Improvisation
Cara|Rogers|Rugby
Cara|Rogers|Debate
Ori|Mejia|Debate
sqlite> SELECT first_name, last_name FROM student JOIN student_to_club ON student_to_club.student_id = student.id JOIN club ON student_to_club.club_id = club.id WHERE name = 'Chess'LIMIT 5;
Cara|Rogers
Wing|Gordon
Eagan|Hogan
Jael|Craig
Joshua|Forbes
sqlite> CREATE VIEW student_clubs AS SELECT first_name, last_name, name FROM student JOIN student_to_club ON student_to_club.student_id = student.id JOIN club on student_to_club.club_id=club.id;
sqlite> select * from student_clubs LIMIT 5;
Cara|Rogers|Chess
Cara|Rogers|Improvisation
Cara|Rogers|Rugby
Cara|Rogers|Debate
Ori|Mejia|Debate
sqlite> INSERT INTO student(first_name, last_name, status_id) VALUES ('Egon', 'Spengler', 4);
sqlite> UPDATE student SET status_id = (SELECT id FROM status WHERE label = 'Freshman') WHERE id = 101;
sqlite> SELECT * from student WHERE id = 101;
101|Egon|Spengler|3
Bash Command
jessica@mac Student database solution % echo "SELECT * FROM student LIMIT 5;" | sqlite3 -header -separator ' ' student_data.sqlite
id first_name last_name status_id
1 Cara Rogers 1
2 Ori Mejia 2
3 Leandra Stevens 3
4 Danielle Moody 1
5 Josiah Barber 1
Wednesday, June 23, 2010
Blanton SDSS Data Talk
Blanton gave a detailed talk about SDSS Imaging Data which I think has a bunch of useful information so I want to re-post it here. The pdf of the talk is on the repository. Click on any of the below slides to enlarge.






































Tuesday, June 22, 2010
Introduction to Databases
Add on the Firefox Database Manager: SQLight Manager.
Start by creating a schema for your data.
Every table within your database will have an ID as the first column of your table. This should be the primary key for that table.
You should think about how different tables map to each other and if each column of your table should have unique values or not.
Some documentation is here: http://www.postgresql.org/docs/8.4/interactive/datatype-numeric.html
Start by creating a schema for your data.
Every table within your database will have an ID as the first column of your table. This should be the primary key for that table.
You should think about how different tables map to each other and if each column of your table should have unique values or not.
Some documentation is here: http://www.postgresql.org/docs/8.4/interactive/datatype-numeric.html
SciCoder Repository
The sciCoder repository is here: http://subversion.assembla.com/svn/scicoder
It has the lectures for the course.
It has the lectures for the course.
Monday, June 21, 2010
Environment Variables
Some commands to remember about Environment Variables:
echo $HOME
/Users/jessica
echo $PATH
/Library/Frameworks/Python.framework/Versions/Current/bin:/sw/bin:/sw/sbin:/Library/Frameworks/Python.framework/Versions/Current/bin:/Users/jessica/python/MyModules:/Users/jessica/idl/photoop/bin:/Users/jessica/idl/idlspec2d/bin:/Users/jessica/idl/idlutils/bin:/Applications/rsi/idl/bin:/usr/local/bin:/usr/X11R6/bin:/bin:/sbin:/usr/bin:/usr/sbin:/usr/texbin:/usr/local/bin:/Users/jessica/bin:/usr/local/bin:.:/Users/jessica/bin:/usr/local/bin:.
which ls
/bin/ls
env
MANPATH=/sw/share/man:/Library/Frameworks/Python.framework/Versions/Current/share/man:/usr/X11R6/share/man:/usr/share/man:/usr/X11/man:/sw/lib/perl5/5.10.0/man:/usr/X11R6/man
IDLSPEC2D_DIR=/Users/jessica/idl/idlspec2d
TERM_PROGRAM=Apple_Terminal
TERM=xterm-color
SHELL=/bin/bash
TMPDIR=/var/folders/ay/ay3fH9RRFLSKHrn6zK-vhE+++TI/-Tmp-/
PERL5LIB=/sw/lib/perl5:/sw/lib/perl5/darwin
IDL_DIR=/Applications/rsi/idl
Apple_PubSub_Socket_Render=/tmp/launch-BDx2kp/Render
CVSROOT=:pserver:anonymous@sdsscvs.astro.princeton.edu:/usr/local/cvsroot
TERM_PROGRAM_VERSION=273
KCORRECT_DIR=/Users/jessica/idl/kcorrect
SGML_CATALOG_FILES=/sw/etc/sgml/catalog
USER=jessica
LD_LIBRARY_PATH=:/Users/jessica/idl/kcorrect/lib
COMMAND_MODE=unix2003
SSH_AUTH_SOCK=/tmp/launch-k6t2m6/Listeners
__CF_USER_TEXT_ENCODING=0x1F5:0:0
PHOTOOP_DIR=/Users/jessica/idl/photoop
PATH=/Library/Frameworks/Python.framework/Versions/Current/bin:/sw/bin:/sw/sbin:/Library/Frameworks/Python.framework/Versions/Current/bin:/Users/jessica/python/MyModules:/Users/jessica/idl/photoop/bin:/Users/jessica/idl/idlspec2d/bin:/Users/jessica/idl/idlutils/bin:/Applications/rsi/idl/bin:/usr/local/bin:/usr/X11R6/bin:/bin:/sbin:/usr/bin:/usr/sbin:/usr/texbin:/usr/local/bin:/Users/jessica/bin:/usr/local/bin:.:/Users/jessica/bin:/usr/local/bin:.
MKL_NUM_THREADS=1
XML_CATALOG_FILES=/sw/etc/xml/catalog
SPECTRO_DATA=/Users/jessica/spectro
PWD=/Users/jessica
LANG=en_US.UTF-8
SHLVL=1
HOME=/Users/jessica
IDL_PATH=+./:+/Users/jessica/Documents/Research/IDLCode/newman/spherepairs:+/Users/jessica/Documents/Research/IDLCode:+/Users/jessica/Documents/Research/IDLCode/newman:+/Users/jessica/Documents/Research/QSOClustering/pro:+/Users/jessica/idl:+/Users/jessica/idl/photoop/pro:+/Users/jessica/idl/idlspec2d/pro:+/Users/jessica/idl/idlutils/pro:+/Users/jessica/idl/idlutils/goddard/pro:+/Applications/rsi/idl/lib
PYTHONPATH=/Users/jessica/python/MyModules:/Users/jessica/idl/photoop/bin:/Users/jessica/idl/idlspec2d/bin:/Users/jessica/idl/idlutils/bin:/Applications/rsi/idl/bin:/usr/local/bin:/usr/X11R6/bin:/bin:/sbin:/usr/bin:/usr/sbin:/usr/texbin:/usr/local/bin
LOGNAME=jessica
CVS_RSH=ssh
IDLUTILS_DIR=/Users/jessica/idl/idlutils
INFOPATH=/sw/share/info:/sw/info:/usr/share/info
DISPLAY=/tmp/launch-cHq9RL/org.x:0
QSO_DIR=/Users/jessica/Documents/Research/QSODir
_=/usr/bin/env
type -a python
python is /Library/Frameworks/Python.framework/Versions/Current/bin/python
python is /Library/Frameworks/Python.framework/Versions/Current/bin/python
python is /usr/local/bin/python
python is /usr/bin/python
python is /usr/local/bin/python
python is /usr/local/bin/python
python is /usr/local/bin/python
export VARIABLE="value"
echo $VARIABLE
value
echo $HOME
/Users/jessica
echo $PATH
/Library/Frameworks/Python.framework/Versions/Current/bin:/sw/bin:/sw/sbin:/Library/Frameworks/Python.framework/Versions/Current/bin:/Users/jessica/python/MyModules:/Users/jessica/idl/photoop/bin:/Users/jessica/idl/idlspec2d/bin:/Users/jessica/idl/idlutils/bin:/Applications/rsi/idl/bin:/usr/local/bin:/usr/X11R6/bin:/bin:/sbin:/usr/bin:/usr/sbin:/usr/texbin:/usr/local/bin:/Users/jessica/bin:/usr/local/bin:.:/Users/jessica/bin:/usr/local/bin:.
which ls
/bin/ls
env
MANPATH=/sw/share/man:/Library/Frameworks/Python.framework/Versions/Current/share/man:/usr/X11R6/share/man:/usr/share/man:/usr/X11/man:/sw/lib/perl5/5.10.0/man:/usr/X11R6/man
IDLSPEC2D_DIR=/Users/jessica/idl/idlspec2d
TERM_PROGRAM=Apple_Terminal
TERM=xterm-color
SHELL=/bin/bash
TMPDIR=/var/folders/ay/ay3fH9RRFLSKHrn6zK-vhE+++TI/-Tmp-/
PERL5LIB=/sw/lib/perl5:/sw/lib/perl5/darwin
IDL_DIR=/Applications/rsi/idl
Apple_PubSub_Socket_Render=/tmp/launch-BDx2kp/Render
CVSROOT=:pserver:anonymous@sdsscvs.astro.princeton.edu:/usr/local/cvsroot
TERM_PROGRAM_VERSION=273
KCORRECT_DIR=/Users/jessica/idl/kcorrect
SGML_CATALOG_FILES=/sw/etc/sgml/catalog
USER=jessica
LD_LIBRARY_PATH=:/Users/jessica/idl/kcorrect/lib
COMMAND_MODE=unix2003
SSH_AUTH_SOCK=/tmp/launch-k6t2m6/Listeners
__CF_USER_TEXT_ENCODING=0x1F5:0:0
PHOTOOP_DIR=/Users/jessica/idl/photoop
PATH=/Library/Frameworks/Python.framework/Versions/Current/bin:/sw/bin:/sw/sbin:/Library/Frameworks/Python.framework/Versions/Current/bin:/Users/jessica/python/MyModules:/Users/jessica/idl/photoop/bin:/Users/jessica/idl/idlspec2d/bin:/Users/jessica/idl/idlutils/bin:/Applications/rsi/idl/bin:/usr/local/bin:/usr/X11R6/bin:/bin:/sbin:/usr/bin:/usr/sbin:/usr/texbin:/usr/local/bin:/Users/jessica/bin:/usr/local/bin:.:/Users/jessica/bin:/usr/local/bin:.
MKL_NUM_THREADS=1
XML_CATALOG_FILES=/sw/etc/xml/catalog
SPECTRO_DATA=/Users/jessica/spectro
PWD=/Users/jessica
LANG=en_US.UTF-8
SHLVL=1
HOME=/Users/jessica
IDL_PATH=+./:+/Users/jessica/Documents/Research/IDLCode/newman/spherepairs:+/Users/jessica/Documents/Research/IDLCode:+/Users/jessica/Documents/Research/IDLCode/newman:+/Users/jessica/Documents/Research/QSOClustering/pro:+/Users/jessica/idl:+/Users/jessica/idl/photoop/pro:+/Users/jessica/idl/idlspec2d/pro:+/Users/jessica/idl/idlutils/pro:+/Users/jessica/idl/idlutils/goddard/pro:+/Applications/rsi/idl/lib
PYTHONPATH=/Users/jessica/python/MyModules:/Users/jessica/idl/photoop/bin:/Users/jessica/idl/idlspec2d/bin:/Users/jessica/idl/idlutils/bin:/Applications/rsi/idl/bin:/usr/local/bin:/usr/X11R6/bin:/bin:/sbin:/usr/bin:/usr/sbin:/usr/texbin:/usr/local/bin
LOGNAME=jessica
CVS_RSH=ssh
IDLUTILS_DIR=/Users/jessica/idl/idlutils
INFOPATH=/sw/share/info:/sw/info:/usr/share/info
DISPLAY=/tmp/launch-cHq9RL/org.x:0
QSO_DIR=/Users/jessica/Documents/Research/QSODir
_=/usr/bin/env
type -a python
python is /Library/Frameworks/Python.framework/Versions/Current/bin/python
python is /Library/Frameworks/Python.framework/Versions/Current/bin/python
python is /usr/local/bin/python
python is /usr/bin/python
python is /usr/local/bin/python
python is /usr/local/bin/python
python is /usr/local/bin/python
export VARIABLE="value"
echo $VARIABLE
value
Friday, June 18, 2010
Princeton Progress
We have some confusing findings from this week. First of all, I re-ran the reconstruction on a smaller angular scale (as both Alexia and Erin told me to do as I was going out to way to large of scales) and got the following for the angular correlation functions:
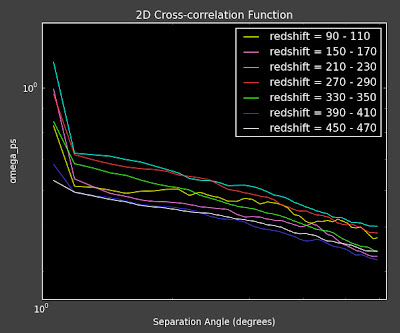
This concerns Alexia and me because first of all they are almost featureless on this degree scale (they look really flat). Second, they seem to move in the wrong direction as a function of redshift bin. We would expect that as we go out further in redshift, the same angular separation would correspond to larger physical separation of objects, and thus the angular correlation should decrease as redshift increases (see below), but we are seeing the opposite.
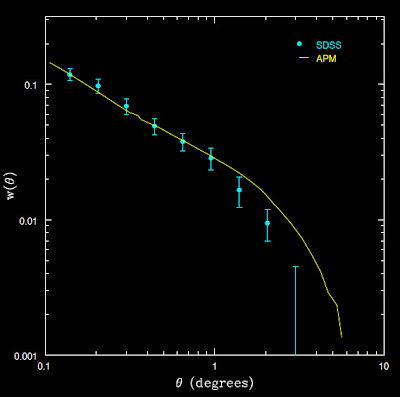 From Connolly et al. arXiv:astro-ph/0107417v2
From Connolly et al. arXiv:astro-ph/0107417v2
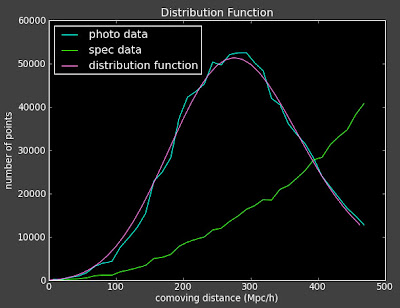

This concerns Alexia and me because first of all they are almost featureless on this degree scale (they look really flat). Second, they seem to move in the wrong direction as a function of redshift bin. We would expect that as we go out further in redshift, the same angular separation would correspond to larger physical separation of objects, and thus the angular correlation should decrease as redshift increases (see below), but we are seeing the opposite.
 From Connolly et al. arXiv:astro-ph/0107417v2
From Connolly et al. arXiv:astro-ph/0107417v2 Here are the 3D correlation functions:

And the best fit reconstruction, which isn't very good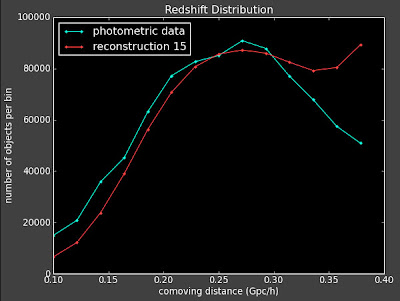

And I am still getting problems with changing the binning dramatically influencing the reconstruction.
List of things to do:
1. Run my angular correlation function on the same data set as in this paper, and see if I get the same solution.
2. Alexia is running the reconstruction on her data sets to see if she gets the same strange behavior for the correlation functions as a function of redshift.
3. Alexia is going to play with the binning and see if she gets the same strange behavior.
4. I need to come up with another way to do the reconstruction that doesn't involve this lambda/tolerance but minimizes the ximat...
List of things to do:
1. Run my angular correlation function on the same data set as in this paper, and see if I get the same solution.
2. Alexia is running the reconstruction on her data sets to see if she gets the same strange behavior for the correlation functions as a function of redshift.
3. Alexia is going to play with the binning and see if she gets the same strange behavior.
4. I need to come up with another way to do the reconstruction that doesn't involve this lambda/tolerance but minimizes the ximat...
Labels:
2D,
Alexia Schulz,
correlation function,
Erin Sheldon,
reconstruction,
redshift
Tuesday, June 15, 2010
Deconstructing Reconstruction
Sorry for the lack of postings lately. I'm at Princeton working with Alexia this week. Hoping to make a lot of progress on the reconstruction project. Remember way back when we were having troubles with binning? Well I am finally trying to get to the bottom of it.
Using my best working run from back in February (man on man has it been that long?) Here is the reconstruction.... (code to reproduce these plots is in the following log file ../logs/100615log.py)
A few things we discovered... first. I wasn't interpolating correctly in getximat. I needed to set interpolate=1. Second, the sximat has some pretty outrageous values in it. This is mostly due to the fact that I am running the reconstruction on too small of angles/separations where I don't have enough objects to have a valid correlation function.
In places where I do have enough objects the interpolation now looks pretty good:
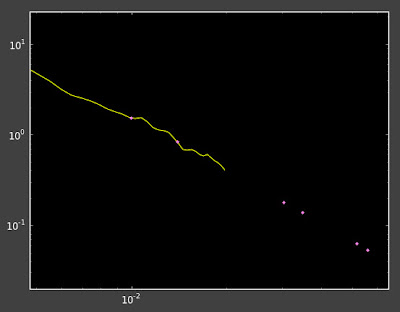 Yellow line is the correlation function, pink dots are the interpolation
Yellow line is the correlation function, pink dots are the interpolation
Using my best working run from back in February (man on man has it been that long?) Here is the reconstruction.... (code to reproduce these plots is in the following log file ../logs/100615log.py)
A few things we discovered... first. I wasn't interpolating correctly in getximat. I needed to set interpolate=1. Second, the sximat has some pretty outrageous values in it. This is mostly due to the fact that I am running the reconstruction on too small of angles/separations where I don't have enough objects to have a valid correlation function.
In places where I do have enough objects the interpolation now looks pretty good:
 Yellow line is the correlation function, pink dots are the interpolation
Yellow line is the correlation function, pink dots are the interpolationThe next thing we discovered is that when I include correlation function in the reconstruction that do not have enough objects in them such that they give sensible answers -- this happens when I try to reconstruct at low redshifts -- my ximat blows up and gives really large values which mess up the interpolation.
So, I've fixed these two problems by
1) Setting interpolation = 1 (as default)
2) Re-running the reconstruction such that it doesn't include low redshifts.
So, I've fixed these two problems by
1) Setting interpolation = 1 (as default)
2) Re-running the reconstruction such that it doesn't include low redshifts.
Labels:
Alexia Schulz,
binning,
interpolation,
reconstruction,
ximat
Wednesday, June 9, 2010
Miscellaneous IDL Commands
qselect -u jessica -s Q | xargs qdel
window,xsize=700,ysize=600
colorobject = qsos.z_tot
xobject = ugcoaddcolor
yobject = grcoaddcolor
thiscolorbin = 0.2
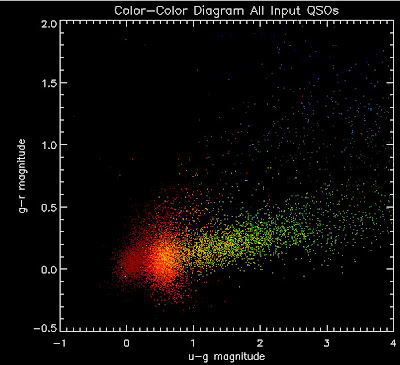
thiscolorstart = 2.0
xtitle = 'u-g magnitude'
ytitle = 'g-r magnitude'
mtitle = 'Color-Color Diagram All Input QSOs'
tempplot, colorobject, xobject, yobject, thiscolorbin, thiscolorstart, xtitle, ytitle, mtitle
data = qsos.z_tot
datamin = 0.0
datamax = 5.0
binsize = 0.1
xtitle = 'Redshift Bin'
ytitle = 'QSOs in Bin'
mtitle = 'Redshift Distribution'
.com histplot.pro
histplot, data, datamin, datamax, binsize, xtitle, ytitle, mtitle
window,xsize=700,ysize=600
colorobject = qsos.z_tot
xobject = ugcoaddcolor
yobject = grcoaddcolor
thiscolorbin = 0.2
thiscolorstart = 2.0
xtitle = 'u-g magnitude'
ytitle = 'g-r magnitude'
mtitle = 'Color-Color Diagram All Input QSOs'
tempplot, colorobject, xobject, yobject, thiscolorbin, thiscolorstart, xtitle, ytitle, mtitle
data = qsos.z_tot
datamin = 0.0
datamax = 5.0
binsize = 0.1
xtitle = 'Redshift Bin'
ytitle = 'QSOs in Bin'
mtitle = 'Redshift Distribution'
.com histplot.pro
histplot, data, datamin, datamax, binsize, xtitle, ytitle, mtitle
Wednesday, June 2, 2010
More likelihood testing
The next test I am running the likelihood with the BOSS QSO's included (I did this before, but it was when I was setting the thresholds wrong, so I'm trying it again).
1. First I make a catalog of all the BOSS and otherwise known QSOs which aren't already in the inputs for which we also have coadded fluxes by spherematching them with the varcat files. The code to do this is in the following log file ../logs/100602_2log.pro. This makes a file: ~/boss/coaddedALLQSOs.fits which has the coadded fluxes (from varcat files) and redshifts of these QSOs. These fluxes are not de-reddened. I've also added the ra/dec to this structure because Schlegel just reminded me of a function of the likelihood_compute that excludes any object in the QSO Catalog when doing the calculation of the likelihoods that has the same ra/dec as the test object. There are 6,063 QSOs in this file.
1. First I make a catalog of all the BOSS and otherwise known QSOs which aren't already in the inputs for which we also have coadded fluxes by spherematching them with the varcat files. The code to do this is in the following log file ../logs/100602_2log.pro. This makes a file: ~/boss/coaddedALLQSOs.fits which has the coadded fluxes (from varcat files) and redshifts of these QSOs. These fluxes are not de-reddened. I've also added the ra/dec to this structure because Schlegel just reminded me of a function of the likelihood_compute that excludes any object in the QSO Catalog when doing the calculation of the likelihoods that has the same ra/dec as the test object. There are 6,063 QSOs in this file.
Here is a plot of these quasars
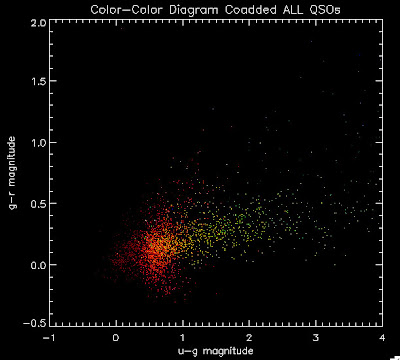
2. Combine this BOSS catalog with SDSS DR5 quasars that are included in the old version of the Monte Carlo. This is done by running Joe Hennawi's qso_photosamp.pro program (to get the SDSS DR5 QSOs), reading in coaddedBOSSQSOs file, de-reddening them both, and then merging them into one file, which is then saved as ~/boss/allbrightQSOMCInput.fits. The code to do this is in the following log file ../logs/100602_3log.pro. (I tried to do this with the SDSS DR7 and was having some problems converting from magnitudes to psffluxes)
3. Change qso_fakephoto.pro to read in the ~/boss/allbrightQSOMCInput.fits file instead of just running qso_photosamp.pro. Save this modified program as ../likelihood/qsocatalog/qso_fakephoto_jess.pro
4. Change hiz_kde_numerator.pro to call qso_fakephoto_jess.pro, instead of qso_fakephoto.pro and save as ../likelihood/qsocatalog/hiz_kde_numerator_jess.pro
5. Follow instructions from this post for how to change/create the luminosity function.
6. Enter the command :
.run hiz_kde_numerator_jess.pro into IDL to generate QSO Catalog.
7. Change likelihood_compute.pro so that it points to this QSO Catalog that you just generated as the input QSO Catalog:
; Read in the QSO
filefile2 = '/home/jessica/repository/ccpzcalib/Jessica/likelihood/qsocatalog/QSOCatalog-Mon-Jun-7-13:56:32-2010.fits'

2. Combine this BOSS catalog with SDSS DR5 quasars that are included in the old version of the Monte Carlo. This is done by running Joe Hennawi's qso_photosamp.pro program (to get the SDSS DR5 QSOs), reading in coaddedBOSSQSOs file, de-reddening them both, and then merging them into one file, which is then saved as ~/boss/allbrightQSOMCInput.fits. The code to do this is in the following log file ../logs/100602_3log.pro. (I tried to do this with the SDSS DR7 and was having some problems converting from magnitudes to psffluxes)
allbrightQSOMCInput.fits QSOs

3. Change qso_fakephoto.pro to read in the ~/boss/allbrightQSOMCInput.fits file instead of just running qso_photosamp.pro. Save this modified program as ../likelihood/qsocatalog/qso_fakephoto_jess.pro
4. Change hiz_kde_numerator.pro to call qso_fakephoto_jess.pro, instead of qso_fakephoto.pro and save as ../likelihood/qsocatalog/hiz_kde_numerator_jess.pro
5. Follow instructions from this post for how to change/create the luminosity function.
6. Enter the command :
.run hiz_kde_numerator_jess.pro into IDL to generate QSO Catalog.
7. Change likelihood_compute.pro so that it points to this QSO Catalog that you just generated as the input QSO Catalog:
; Read in the QSO
filefile2 = '/home/jessica/repository/ccpzcalib/Jessica/likelihood/qsocatalog/QSOCatalog-Mon-Jun-7-13:56:32-2010.fits'
QSO Catalog (including the BOSS quasars)
Subscribe to:
Posts (Atom)



